
April 13, 2025—A collaborative study led by Prof. Xiaojie Lu’s group at the Shanghai Institute of Materia Medica (Chinese Academy of Sciences), Prof. Lu Zhou’s team at Fudan University, and Prof. Yi Sun’s team from the Cancer Institute of Zhejiang University School of Medicine/Translational Medicine Institute of Zhejiang University was published in the international journal Angewandte Chemie International Edition under the title “Proteome-Wide Data Guides the Discovery of Lysine-Targeting Covalent Inhibitors Using DNA-Encoded Chemical Libraries”.
Covalent drugs achieve sustained modulation of target proteins by forming covalent bonds with specific amino acid residues, representing a pivotal direction in modern drug discovery. Current covalent drugs predominantly target cysteine residues near active sites; however, this strategy is limited by the low abundance of cysteine, rendering it inapplicable to protein targets lacking this residue. In prior work, Prof. Zhou’s team established the ABPP-CoDEL strategy, integrating Activity-Based Protein Profiling (ABPP) with Covalent DNA-Encoded Chemical Libraries (CoDELs), to discover a series of tyrosine-targeting covalent inhibitors (J. Am. Chem. Soc. 2023, 145, 25283–25292), expanding the range of targetable amino acids. Compared to cysteine and tyrosine, lysine is more abundant, offering broader potential for overcoming amino acid scarcity and diversifying targetable proteins. Nevertheless, a systematic CoDEL screening platform dedicated to lysine remained absent.
In this study, the team constructed a protein dataset containing highly reactive and ligandable lysine sites by integrating compound- and warhead-driven ABPP data, providing theoretical support for target selection. Subsequently, researchers introduced eight lysine-targeting covalent warheads with distinct reaction mechanisms and synthesized a CoDEL library comprising 10.7 million compounds. Covalent screening identified lysine-targeting inhibitors against PGAM1, BRD family proteins, and UBE2N. Key findings include:
Compound 1 acts as a photoactive covalent ligand for PGAM1;
Compound 4 forms reversible covalent bonds with unexplored sites in the bromodomains of BRD family proteins;
Compound 9 irreversibly binds to UBE2N, inducing conformational changes in the UBE2N/UBE2V2 complex, disrupting ubiquitin chain formation, and interfering with downstream functional activity. This provides a novel mechanism for regulating UBE2N-mediated ubiquitination pathways.
By integrating proteomic data with covalent DEL technology, this study establishes an efficient screening platform for discovering lysine-targeting covalent inhibitors.
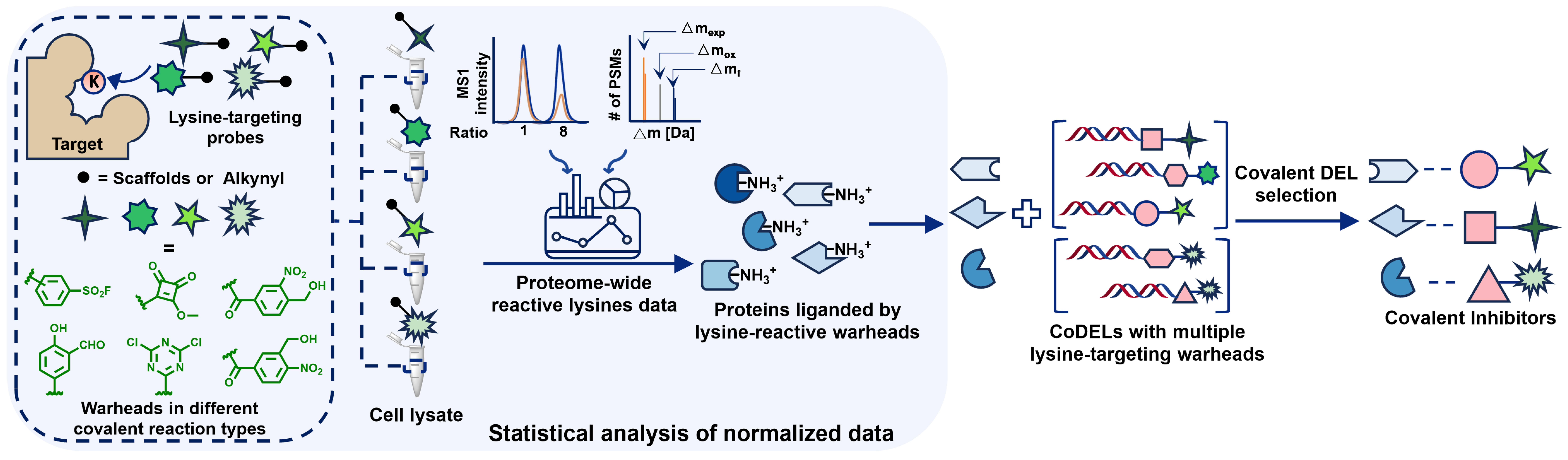
Proteomic Data-Guided Lysine-Targeting CoDEL Screening Strategy
Authors: Xinyuan Wu (Shanghai Institute of Materia Medica), Shunyao Li (Fudan University), and Ting Liang (Zhejiang University) are co-first authors. Corresponding authors are Prof. Xiaojie Lu (Shanghai Institute of Materia Medica), Prof. Lu Zhou (Fudan University), and Prof. Yi Sun (Zhejiang University).
Funding: This work was supported by the Shanghai Synchrotron Radiation Facility, the Mass Spectrometry Platform at Shanghai Institute of Materia Medica, and grants from the National Natural Science Foundation of China, the National Key R&D Program, the Shanghai Municipal Science and Technology Commission, the Self-Research Program of Shanghai Institute of Materia Medica, the Zhejiang Provincial Natural Science Foundation, and the Zhejiang Leading Innovation and Entrepreneurship Team Introduction Program.
Full Article: https://onlinelibrary.wiley.com/doi/10.1002/anie.202505581